Featured Project
The siibra toolsuite — Accessing the EBRAINS atlases interactively and programmatically
date: Dec 01 2021
Let’s introduce you to a dynamic toolsuite for interacting with the EBRAINS brain atlases! siibra (software interfaces for interacting with brain atlases) is a tightly integrated toolsuite which includes siibra-explorer, an interactive 3D atlas viewer, as well as a dedicated programmatic Python client called siibra-python. The aim of siibra is to provide both programmatic and interactive access to the EBRAINS brain atlases, which include the BigBrain and MNI Colin reference spaces, for example, without duplicating software code.
siibra-explorer, the browser-based viewer for visually exploring the EBRAINS atlases, is hosted at https://atlases.ebrains.eu/viewer. It is tightly integrated with the EBRAINS Knowledge Graph, allowing the seamless querying of semantically and spatially anchored datasets. It allows the exploration of the regions of the human, rat, mouse and (currently pre-released) monkey brains at microscopic detail, as well as the discovery of related multimodal data features. The functionalities of the siibra-explorer include (but aren’t limited to):
- Finding, selecting and navigating brain regions
- Accessing different reference templates and maps supported by the EBRAINS atlases
- Viewing relationships between brain regions
- Accessing datasets by their location in the brain and retrieving relevant detailed information from the EBRAINS Knowledge Graph
- Jumping across spatial resolutions to see the same brain regions at different scales
siibra-python is the comprehensive Python client providing access to the EBRAINS atlases and offering an easy and well-structured way to include maps, reference templates, region definitions and linked datasets into reproducible programmatic workflows. The software is maintained on GitHub. It has been designed to allow safe and convenient interaction with brain regions from different parcellations, to provide streamlined access to multimodal data features linked to brain regions, and to perform basic analyses of region-specific data features. The intention is to unify interaction with brain atlas data at different spatial scales, including parcellations and datasets at the millimetre scale in the MNI space, as well as microstructural maps and microscopic data in the BigBrain space.
Check out the siibra toolsuite yourself under the following links. We’d also be delighted to have you participate in our discussions on GitHub!
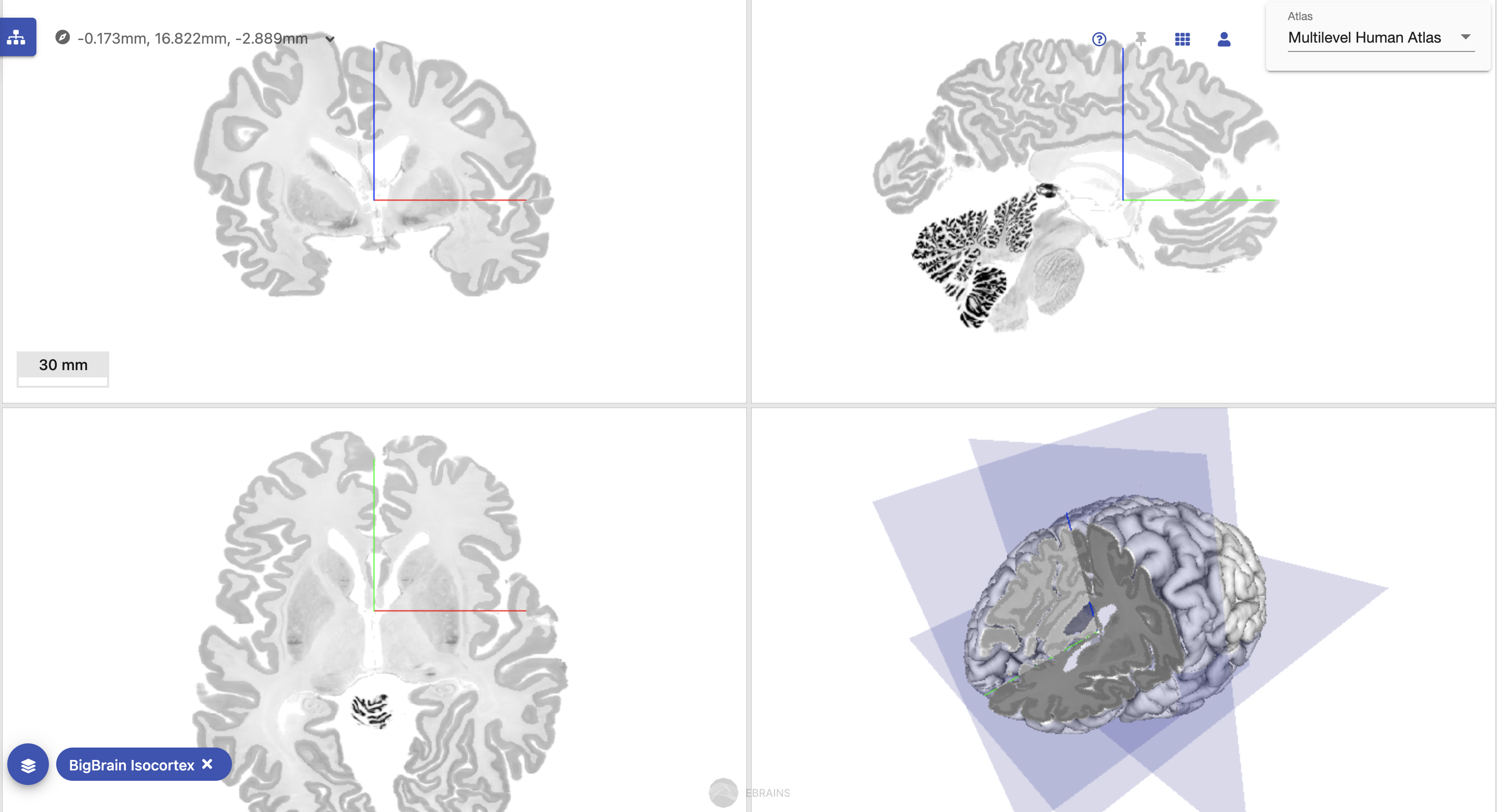
Exploring the BigBrain reference space with the Multilevel Human Brain Atlas using siibra-explorer.
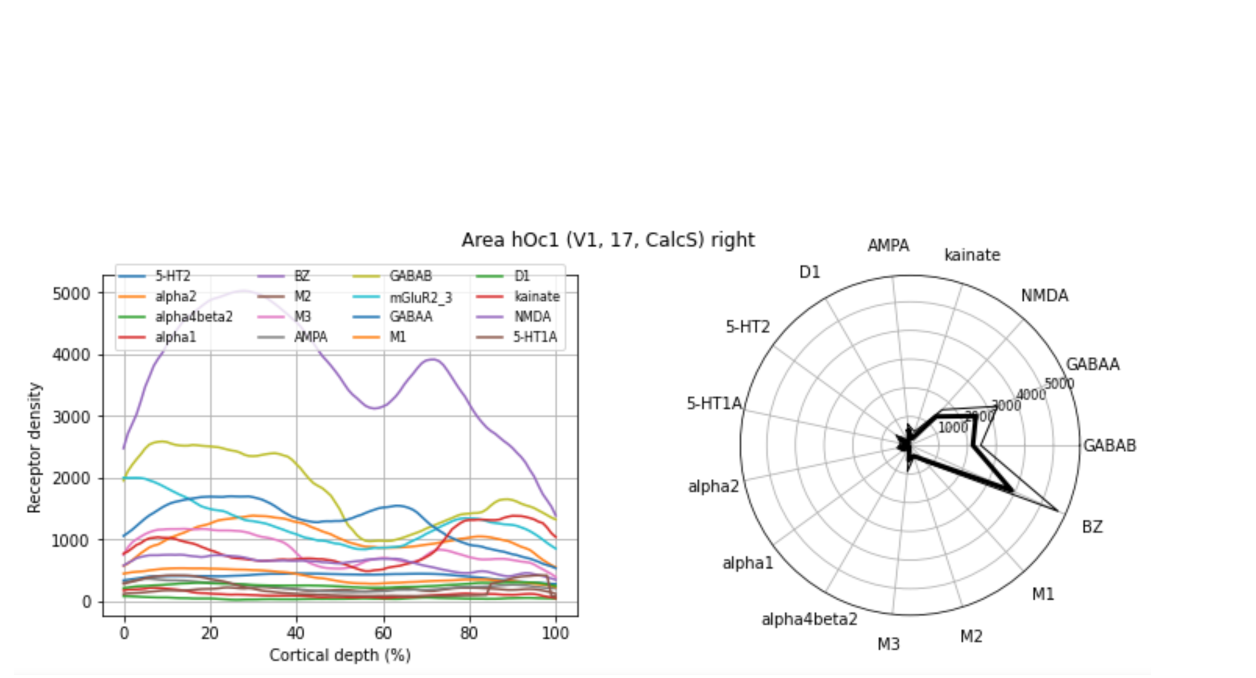
Finding and retrieving neurotransmitter receptor measurements for the primary visual cortex using siibra-python
import siibra
atlas = siibra.atlases['human']
v1 = atlas.get_region('v1',parcellation="julich 2.9")
r, = siibra.get_features(v1,"receptor")
fig = r.plot(r.matched_region.name)
#copy this code to create the graphs