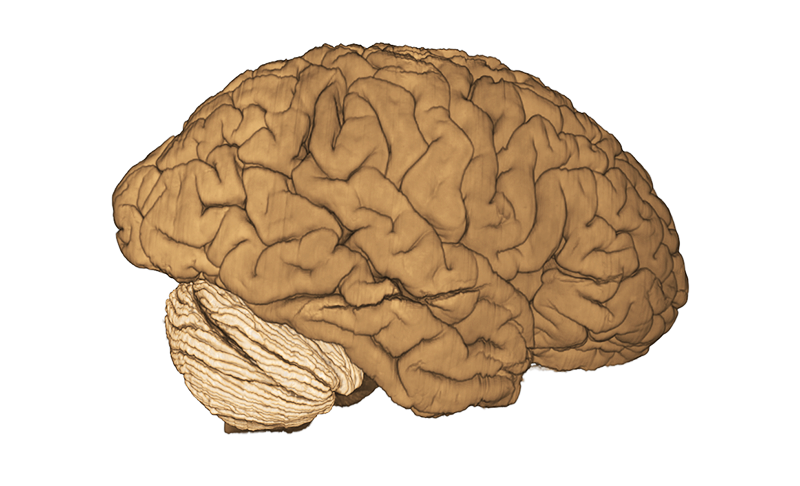
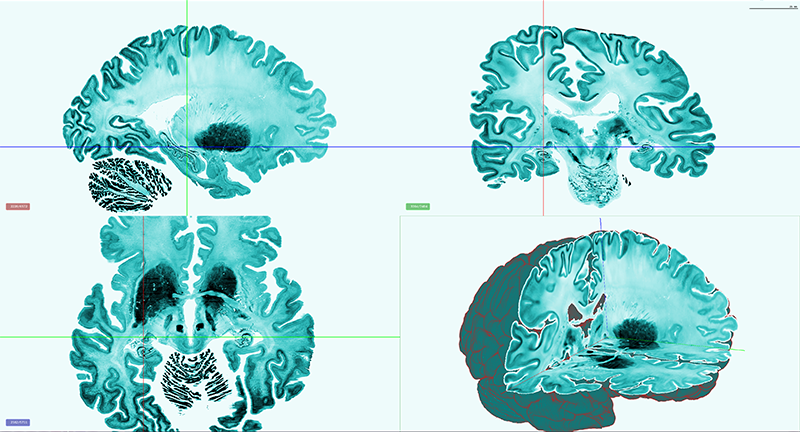
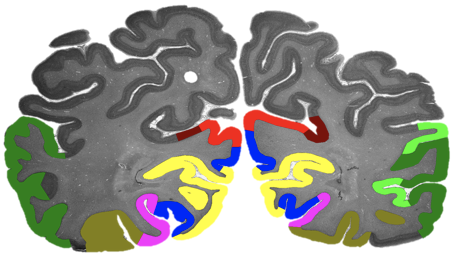
BigBrain Goals
 Building a highly detailed multimodal atlas
Building a highly detailed multimodal atlas
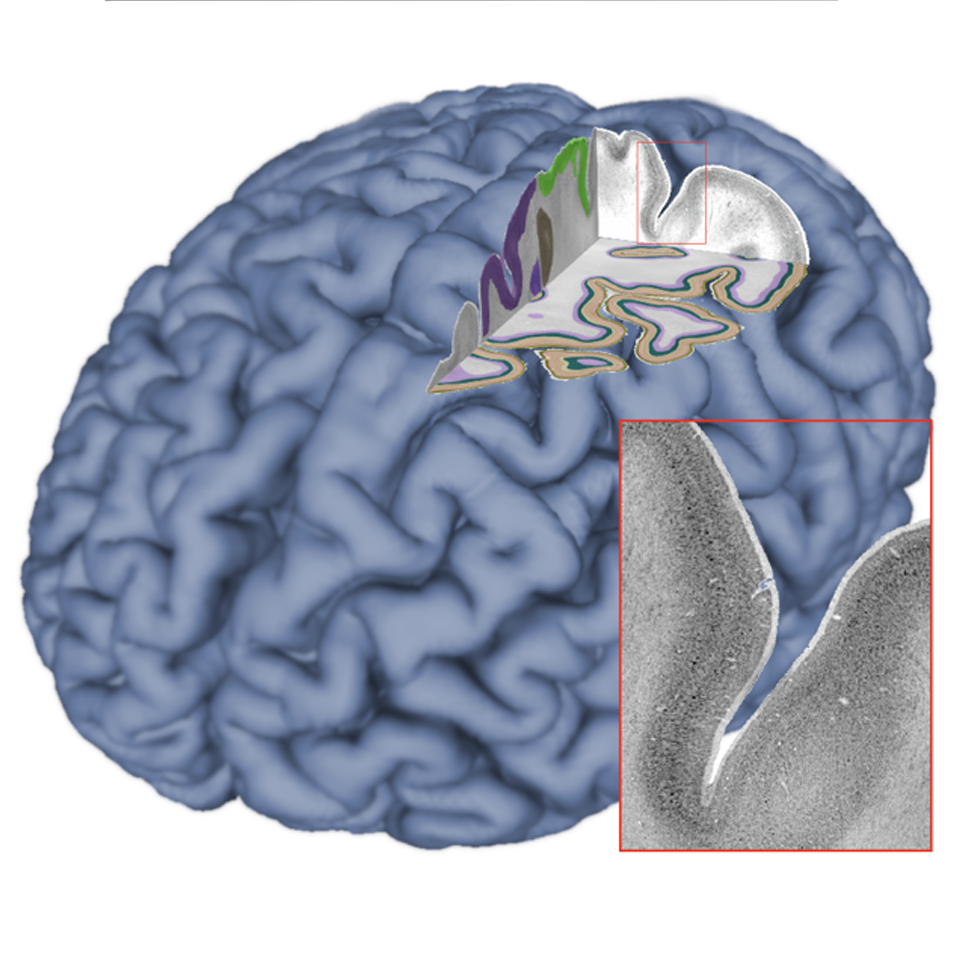
By developing novel AI-based methods for analyzing high-resolution image data, we create highly detailed maps of brain areas, cortical layers, and subcortical structures for our microscopic brain models. We complement the cytoarchitectonic model by modalities that cover fibre- and chemoarchitecture, working towards a multimodal characterization of the human brain at the microscopic scale. Furthermore, we work towards a model with a resolution in 1 micron range, which resolves individual neuronal cell bodies.
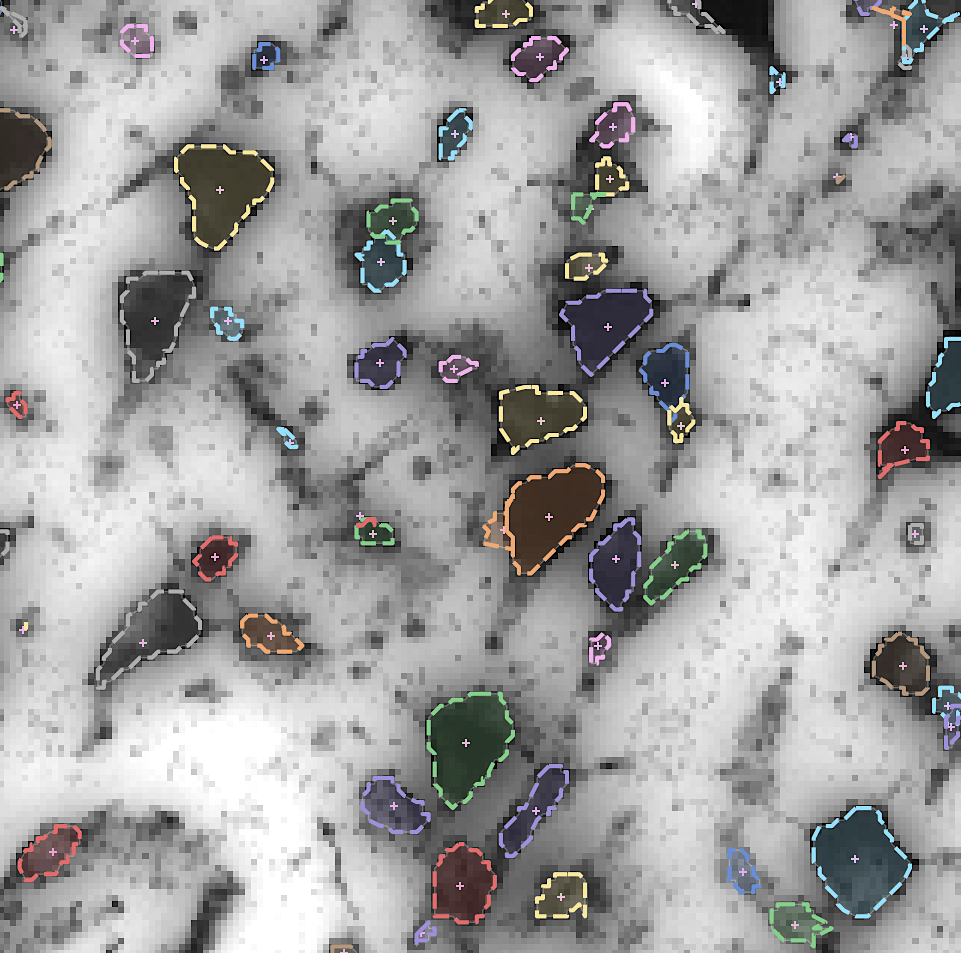 Build on the existing 3D BigBrain as a microscopic reference template
Build on the existing 3D BigBrain as a microscopic reference template
We integrate our ongoing developments with the original BigBrain model (Amunts, Evans et al. 2013) in order to foster a common microscopic reference template. This includes efforts on careful spatial registration across subjects and modalities, and propagation of maps of brain regions.
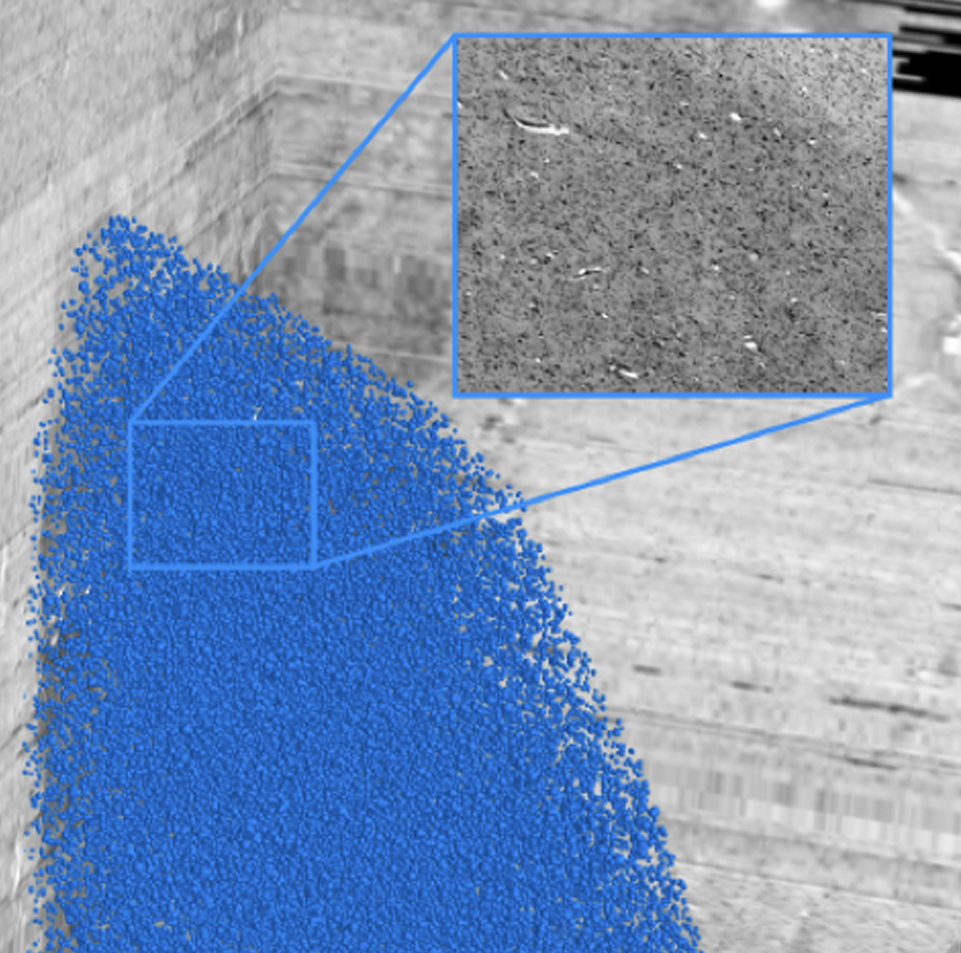 Building a distributed platform for big neuroscience data
Building a distributed platform for big neuroscience data
By increasing the resolution to the level of individual neuronal cell bodies, the size of whole brain models reaches the Petabyte scale. To address such Big Data challenge, we establish specific workflows for distributed data management and processing, and tools for remote visualization and annotation of 2D and 3D data. This also requires standardization efforts for data and metadata formats. We align these infrastructure development efforts with the developments of the Human Brain Project and the HBHL program to maximize compatibility with the international initiatives.
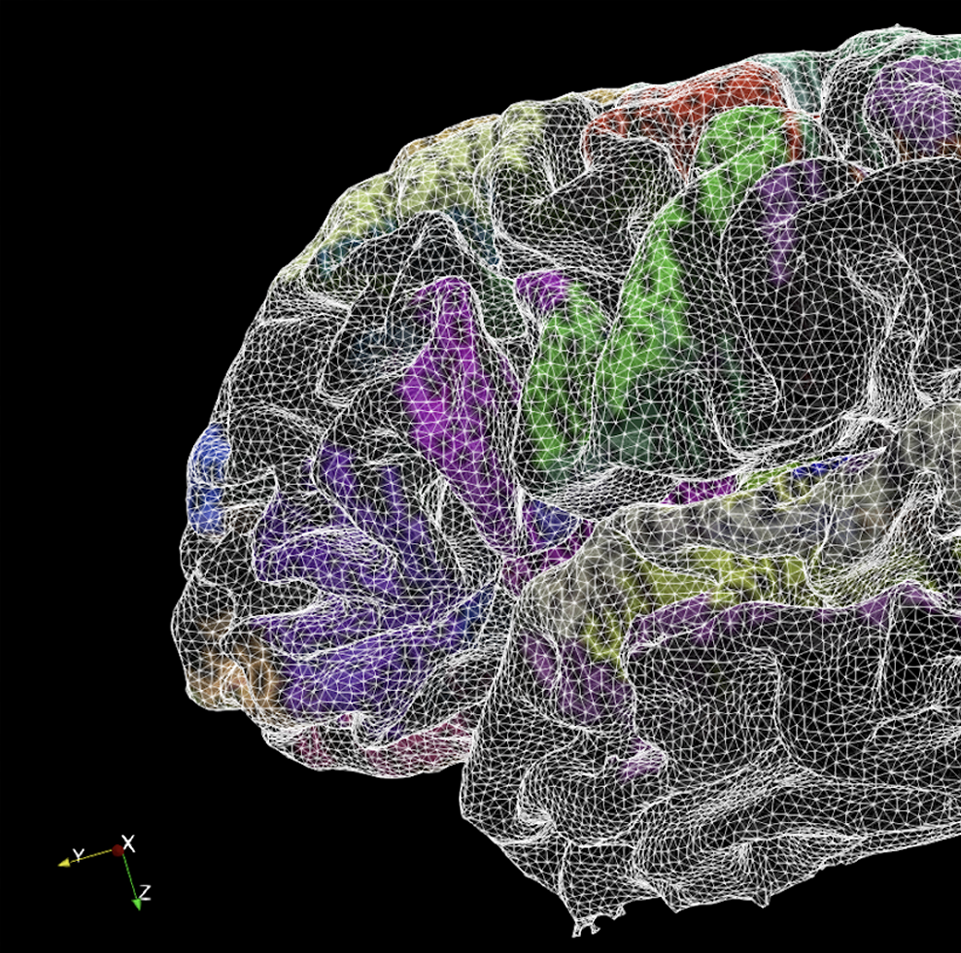 AI link to data science and brain inspired computing
AI link to data science and brain inspired computing
The BigBrain project is entangled with AI research in two fundamental ways - by developing novel image analysis methods based on Machine and Deep Learning, and by contributing knowledge about the microstructural organization of biological neural networks in the brain. The project initiates early cooperation with researchers in brain inspired AI and computing to incorporate information about the human brain into the design of artificial systems.
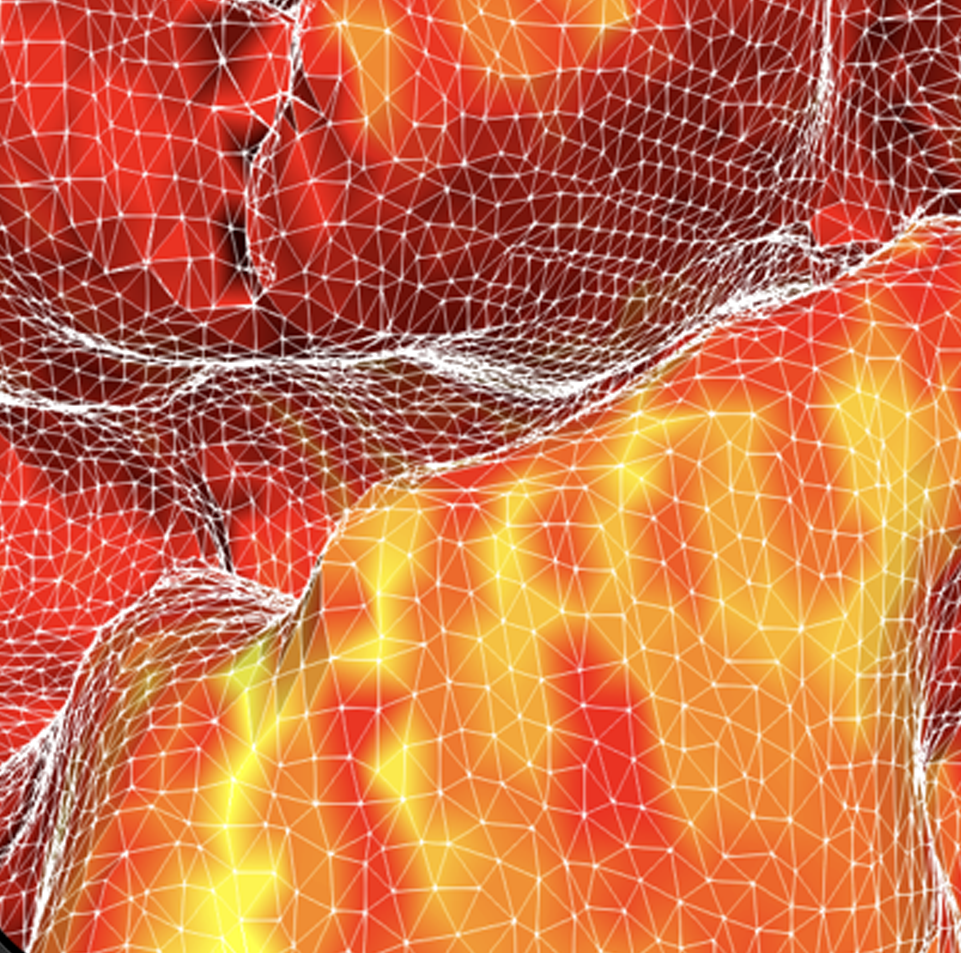 Virtual BigBrain and simulation platform
Virtual BigBrain and simulation platform
The VirtualBrain (TVB) uses empirical knowledge of brain structure to constrain models that simulate network dynamics, which in turn can be validated with experimental observations of brain function. Complementing related work in the Human Brain Project, we support the development of TVB models with increased spatial resolution and regional heterogeneity by informing them with microscopic resolution data, and evaluating optimized software and hardware environments for the CBRAIN platform.
 Open Community Resource
Open Community Resource
The BigBrain Project continues to build an open resource and a lively scientific community within a collaborative research and training environment. We will share our datasets and tools through the platform, aim to integrate them with the EBRAINS and CBRAIN ecosystems, and engage with the BigBrain user and contributor community through open project meetings, workshops, and online services.