Featured Project
Neuromaps – structural and functional interpretation of brain maps
date: July 11 2022
Neuromaps is an open-source Python toolbox for accessing, transforming, and analyzing structural and functional brain annotations. Neuromaps is well situated for contextualizing BigBrain data, such as cytoarchitectonic layer intensities, to the broader structural and functional architecture of the brain. Additionally, neuromaps can be used alongside BigBrain data to better inform computational models of the brain, facilitating cross-modality multi-scale research.
Library
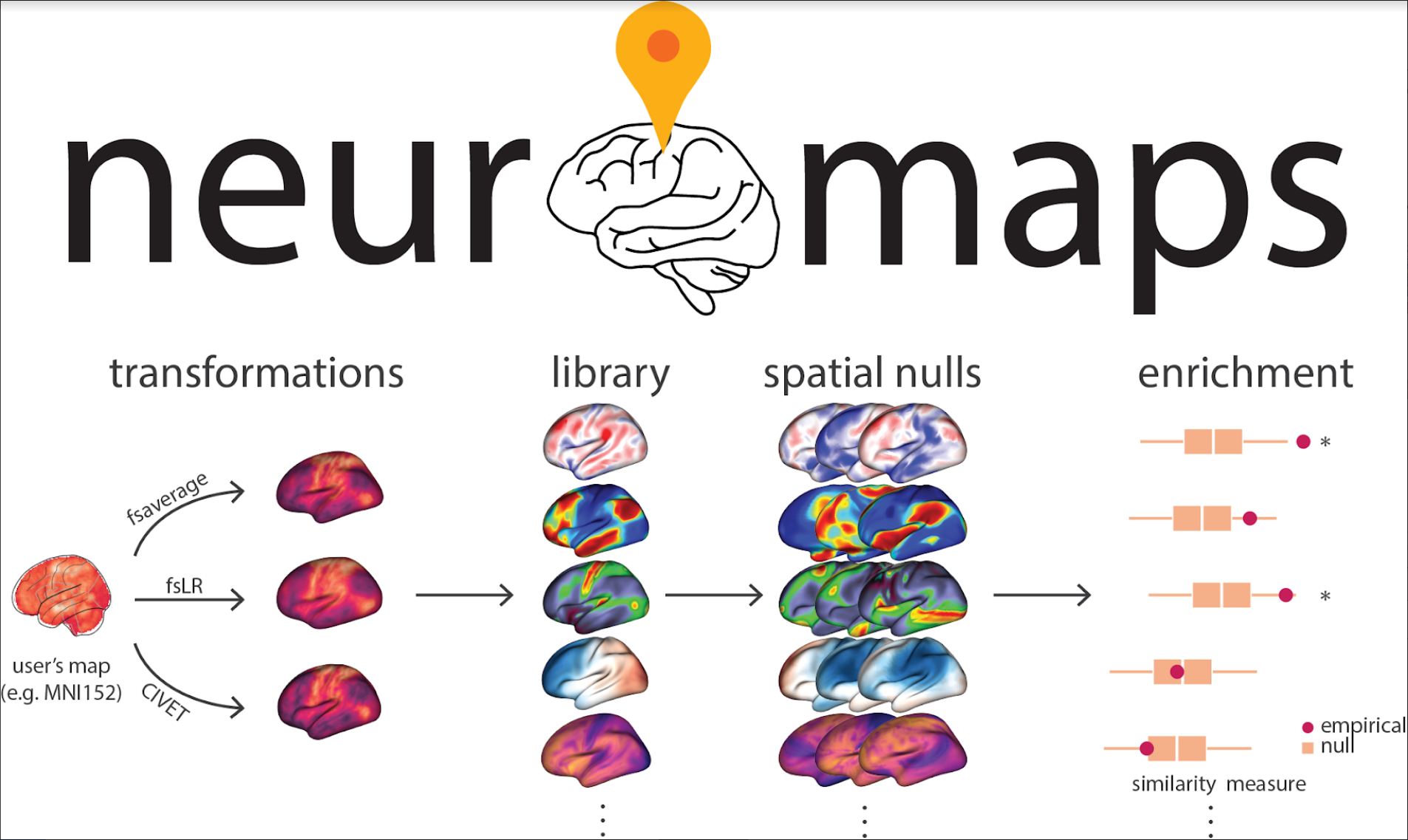
The current instance of neuromaps hosts over 40 volumetric and surface-based brain annotations from a range of imaging modalities - including maps of gene expression, neurotransmitter receptors, metabolism, neurophysiological oscillations, developmental and evolutionary expansion, functional hierarchy, individual functional variability, and cognitive specialization - and will continue to expand through its integrated contribution pipeline.
Transformations
A common challenge when relating novel neuroimaging data to the broader literature is finding a common coordinate space or parcellation in which to conduct the analyses. The neuromaps toolbox provides robust transformation functionality between four standard coordinate systems: MNI-152, fsaverage, fsLR, and CIVET. Bonus: neuromaps can also parcellate!
Spatial nulls
The primary goal for transforming brain maps to a common coordinate system is to statistically compare their spatial topographies. The neuromaps software package implements nine spatial null model frameworks that allow for statistical comparison between brain maps while accounting for spatial autocorrelation. These null models can be used on surface, volumetric, or parcellated data.
Get started
- Read our preprint (or tweeprint)
- Access our code on Github
- See our documentation